PostgreSQL for AI: Storing and Searching Embeddings with pgvector
Mar 10, 2025
Summary
Vector databases are essential for modern AI applications like semantic search, recommendation systems, and natural language processing. They allow us to store and query high-dimensional vectors efficiently. With the pgvector extension PostgreSQL becomes a powerful vector database, enabling you to combine traditional relational data with vector-based operations.
In this post, we will walk through the full process:
Installing PostgreSQL and pgvector Setting up a vector-enabled database Generating embeddings using Ollama Running similarity queries with Python By the end, you’ll be able to store, query, and compare high-dimensional vectors in PostgreSQL, opening up new possibilities for AI-powered applications.
Installing PostgreSQL
- Download the Installer:
Head to the official PostgreSQL website: postgresql.org/downloads/ Choose your operating system (Windows, macOS, Linux). Download the appropriate installer.
- Run the Installer:
Launch the downloaded installer. Follow the on-screen instructions. Key points during installation:
- Installation Directory: Choose where you want to install Postgres. If you’re going to use a lot of data you want to install postgres on your largest drive. Components: It is recommended to install the PostgreSQL Server, pgAdmin 4, Stack Builder, and Command Line Tools.
- Data Directory: Select where database files will be stored.
- Password: Set a strong password for the “postgres” superuser. Remember this password!
- Port: The default port is 5432. It’s usually best to leave it as is.
- Locale Here: Select your preferred locale. I just used the default.
- Verify the Installation:
pgAdmin: If you installed pgAdmin, open it. You should be able to connect to your PostgreSQL server using the “postgres” user and the password you set.
Installing the pgvector extension
Windows installation
I work on windows. For some AI things it can be challenging to get working.
- Clone the repo: pgvector.
- Compiler: For me I had to install the Visual Studio Community Edition, specifically you need the c compiler and libraries.
- PGROOT You must know where PostgreSQL is installed
- Compile the project
set "PGROOT=C:\Program Files\PostgreSQL\16"
git clone https://github.com/pgvector/pgvector.git
cd pgvector
nmake /F Makefile.win
nmake /F Makefile.win install
Check the console output to ensure everything installed successfully.
Linux
sudo apt install postgresql-16-pgvector
After installation, restart the PostgreSQL service to enable the pgvector extension.
sudo systemctl restart postgresql
You can check the status of the service:
sudo systemctl status postgresql
Mac
brew install postgresql
brew install pgvector
Setting up your first vector database
- Create the schema: Create a new schema using pgAdmin.
In the tree view click on the Login/Group roles Rigth-click -> Create -> Login Group Role…

On the General page enter vectordb

On the Definition page enter a password for this user.

On the Privileges enable the user to login.

- Create the database
Use the tree to ceate the database

Make sure and use the role we created as the owner for this database.

- Enable the pgvector extension
You do this through SQL. So we will open the Query Tool. You can do this from the tree view
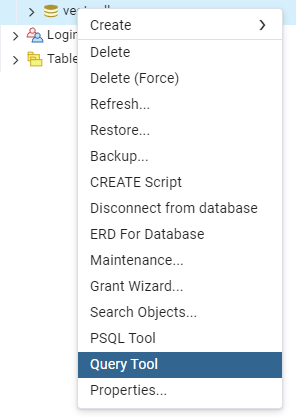
CREATE EXTENSION IF NOT EXISTS vector;
Querying and Comparing Vectors with pgvector in Python
In this section, we’ll explore how to query and compare vectors in pgvector using Python. If you’re looking to use vectors for tasks like recommendation systems, text search, or similarity comparison, Python makes it easy to interface with PostgreSQL and perform vector queries.
For this, we’ll use the popular PostgreSQL adapter for Python, psycopg2.
Setting Up Your Python Environment
To interact with PostgreSQL and use pgvector, you first need to install psycopg2:
pip install psycopg2
I know I am going to use mxbai-embed-large model to generate embeddings so I know this model outputs embeddings of size 1024.
Let’s create this data table:
CREATE TABLE IF NOT EXISTS bible_embeddings (
id SERIAL PRIMARY KEY,
verse TEXT NOT NULL,
text TEXT NOT NULL,
embedding vector(1024) -- Adjust dimension based on your model
);
This table is going to store
- the verse
- the verse’s text
- the generated embeddings for the text from the King James bible
Note: Ensure the embedding dimension matches the model’s output size.
Example 1: Connecting to PostgreSQL
First, establish a connection to your PostgreSQL database using psycopg2:
import os
from dotenv import load_dotenv
import psycopg2
# Load environment variables from .env file
load_dotenv()
def get_connection():
"""Establishes a secure connection using environment variables."""
return psycopg2.connect(
dbname=os.getenv("DB_NAME"),
user=os.getenv("DB_USER"),
password=os.getenv("DB_PASSWORD"),
host=os.getenv("DB_HOST"),
port=os.getenv("DB_PORT")
)
conn = get_connection()
cursor = conn.cursor()
You will need to set env variables I used a .env file.
DB_NAME=your_db
DB_USER=your_username
DB_PASSWORD=your_password
DB_HOST=localhost
DB_PORT=5432
Example 2: Creating the embeddings
Here I will use ollama:
import ollama
import numpy as np
# Function to get embeddings using Ollama
def get_embedding(text):
try:
embedding_data = ollama.embeddings(model="mxbai-embed-large", prompt=text)
embedding_data = embedding_data["embedding"] # Extract embedding
return embedding_data
except Exception as e:
print(f" Unexpected error generating embedding: {e}")
return None # Return None if an error occurs
Example 3: Store the embeddings
This will store a large amount of data with embeddings in your table. I chose the bible it is a medium size book. 31102 verses.
import psycopg2
# Function to insert Bible embeddings into PostgreSQL
def store_bible_embeddings(bible_texts):
print(f" Storing Bible embeddings in PostgreSQL... {len(bible_texts)}")
conn = get_connection()
cursor = conn.cursor()
total = len(bible_texts)
for i, (verse, text) in enumerate(bible_texts, start=1):
embedding = get_embedding(text)
cursor.execute(
"INSERT INTO bible_embeddings (verse, text, embedding) VALUES (%s, %s, %s);",
(verse, text, embedding)
)
if i % 1000 == 0 or i == total:
percent_done = (i / total) * 100
print(f" Inserted {i}/{total} Bible verses ({percent_done:.2f}% done)")
conn.commit()
conn.close()
print(" Bible embeddings stored in PostgreSQL!")
bible_texts = get_bible_text() # Fetches (verse, text)
store_bible_embeddings(bible_texts)
Example 4: Querying Vectors
Now that you have vector data in your database, let’s move on to querying and comparing vectors using similarity measures like Euclidean distance and Cosine similarity.
Querying with Euclidean Distance
You can compare the embedding of an item in your database to a query vector using Euclidean distance (<=> operator in SQL). Here’s how you can do it in Python:
Note: take note of how I built the query for me this worked.
SELECT verse, text, 1 - (embedding <=> '{embedding}') AS similarity
FROM bible_embeddings
ORDER BY embedding <=> '{embedding}'
LIMIT {top_k};
import psycopg2
def find_similar_bible_verses(shakespeare_sentence, top_k=5):
embedding = get_embedding(shakespeare_sentence) # Get embedding as a list
conn = get_connection()
cursor = conn.cursor()
query = """
SELECT verse, text, 1 - (embedding <=> %s) AS similarity
FROM bible_embeddings
ORDER BY embedding <=> %s
LIMIT %s;
"""
cursor.execute(query, (embedding, embedding, top_k))
results = cursor.fetchall()
conn.close()
return results
# Example Query
query_text = "To be, or not to be, that is the question."
bible_matches = find_similar_bible_verses(query_text)
print(f"\n **Bible Verses Similar to Shakespeare's Quote: {query_text}**")
for verse, text, similarity in bible_matches:
print(f"{verse} - {text} (Similarity: {similarity:.4f})")
**Bible Verses Similar to Shakespeare's Quote: To be, or not to be, that is the question.**
Co2 8:12 - For if there be first a willing mind, it is accepted according to that a man hath, and not according to that he hath not.~ (Similarity: 0.6588)
Mat 11:3 - And said unto him, Art thou he that should come, or do we look for another?~ (Similarity: 0.6555)
Kg2 2:10 - And he said, Thou hast asked a hard thing: nevertheless, if thou see me when I am taken from thee, it shall be so unto thee; but if not, it shall not be so.~ (Similarity: 0.6362)
Job 39:9 - Will the unicorn be willing to serve thee, or abide by thy crib?~ (Similarity: 0.6322)
Gal 1:10 - For do I now persuade men, or God? or do I seek to please men? for if I yet pleased men, I should not be the servant of Christ.~ (Similarity: 0.6302)
This query calculates the Euclidean distance between the stored vectors and the query vector using the <=> operator. Results are sorted by similarity, with the closest matches appearing first.
Querying with Cosine Similarity
If you prefer to use Cosine similarity, you can modify the query as follows:
import psycopg2
def find_similar_bible_verses_cosine(shakespeare_sentence, top_k=5):
embedding = get_embedding(shakespeare_sentence) # Get embedding as a list
conn = get_connection()
cursor = conn.cursor()
cursor.execute("""
SELECT verse, text, embedding <=> %s AS similarity
FROM bible_embeddings
ORDER BY similarity DESC
LIMIT %s;
""", (str(embedding), top_k))
# Fetch and print results
results = cursor.fetchall()
return results
query_text = "To be, or not to be, that is the question."
bible_matches = find_similar_bible_verses_cosine(query_text)
for result in bible_matches:
print(f"ID: {result[0]}, Name: {result[1]}, Cosine Similarity: {result[2]}")
In this case, the <=> operator can also be used for cosine similarity. The results are sorted by similarity, where values closer to 1 (for cosine similarity) indicate a higher degree of similarity.
ID: Num 31:10, Name: And they burnt all their cities wherein they dwelt, and all their goodly castles, with fire.~, Cosine Similarity: 0.7576054290461446
ID: Est 9:6, Name: And in Shushan the palace the Jews slew and destroyed five hundred men.~, Cosine Similarity: 0.7569403794042351
ID: Act 18:17, Name: Then all the Greeks took Sosthenes, the chief ruler of the synagogue, and beat him before the judgment seat. And Gallio cared for none of those things.~, Cosine Similarity: 0.7554121530553088
ID: Deu 3:6, Name: And we utterly destroyed them, as we did unto Sihon king of Heshbon, utterly destroying the men, women, and children, of every city.~, Cosine Similarity: 0.750413240364846
ID: Deu 2:34, Name: And we took all his cities at that time, and utterly destroyed the men, and the women, and the little ones, of every city, we left none to remain:~, Cosine Similarity: 0.7503269107409909
Cosine Similarity vs. Euclidean Distance: When to Use Each?
When working with vector search, choosing the right similarity metric is crucial. The two most common options are Cosine Similarity and Euclidean Distance, and each is best suited for different scenarios.
1. Cosine Similarity (Angle-Based Similarity)
Best for: Text embeddings, NLP tasks, recommendation systems
Measures the angle between two vectors, ignoring magnitude.
Returns a similarity score between -1 (opposite) and 1 (identical).
Use when vector magnitude doesn’t matter, e.g., comparing semantic meaning of text.
Example Use Cases:
- Text similarity search (e.g., finding similar documents)
- Recommendation systems (e.g., “users who liked this also liked…”)
- Clustering high-dimensional embeddings
2. Euclidean Distance (Magnitude-Based Distance)
Best for: Geospatial data, image embeddings, numerical feature vectors
Measures the absolute distance between two points in space.
More sensitive to vector magnitude and scale than Cosine Similarity.
Use when absolute differences matter, e.g., finding images with similar pixel distributions.
Example Use Cases:
- Image similarity search (e.g., retrieving similar-looking images)
- Geospatial search (e.g., finding the nearest location)
- Low-dimensional numerical feature comparisons
Quick Rule of Thumb
Use Cosine Similarity when dealing with high-dimensional, unnormalized data (e.g., text embeddings).
Use Euclidean Distance when absolute position and magnitude matter (e.g., images, geospatial data).
Using Indexing to Speed Up Searches
To optimize the vector similarity search for large datasets, you can create an index on the vector column. As mentioned earlier, an IVF (Inverted File Index) is a great option for speeding up queries.
Why Use an IVF Index for pgvector?
When working with high-dimensional vector data, such as embeddings from machine learning models, query performance becomes a major concern. Without indexing, vector similarity searches require a brute-force scan through every row in your database, which is computationally expensive. This is where IVF (Inverted File Index) comes in.
IVF (Inverted File Index) is a technique designed to speed up nearest neighbor search by dividing the dataset into smaller, more manageable groups.
Here’s how it works conceptually:
-
Partitioning the Data (Clustering Stage)
- Before performing queries, IVF clusters the vectors into centroids (groups).
- These clusters are usually generated using k-means clustering.
- Each data point (vector) is assigned to the nearest centroid.
-
Query Execution (Approximate Nearest Neighbor Search)
- When a query vector is provided, instead of scanning all stored vectors, the search first identifies the most relevant clusters.
- Only vectors within the closest clusters are searched, significantly reducing the number of distance calculations.
- This makes retrieval much faster compared to a brute-force search.
Why is IVF Beneficial?
Speeds Up Search: Instead of scanning millions of vectors, it narrows the search to a small subset, reducing query time from seconds to milliseconds.
Scalability: Works well for large datasets where brute-force search becomes impractical.
Customizable Trade-off: You can adjust the number of clusters (k) to balance speed vs. accuracy. More clusters → finer-grained search (but slower).
Supports Different Similarity Metrics: Works with L2 (Euclidean distance), inner product, cosine similarity.
In pgvector, you can create an IVF index as follows:
-- Create an IVF index on the embedding column
CREATE INDEX ON bible_embeddings
USING ivfflat (embedding vector_l2_ops) WITH (lists = 100);
Explanation of Parameters
ivfflat→ This tells PostgreSQL to use an Inverted File Index.vector_l2_ops→ This specifies that L2 (Euclidean distance) should be used for similarity.lists = 100→ The number of clusters (centroids) to use (this value can be tuned for performance).
Tuning lists
- A higher value (e.g.,
1000) increases accuracy but takes longer to query. - A lower value (e.g.,
10) speeds up search but may reduce retrieval quality.
After creating this index, your similarity searches will be faster, especially when dealing with large datasets of vectors.
IVF vs. Brute-Force Search: A Performance Comparison
| Approach | Speed | Accuracy | Best For |
|---|---|---|---|
Brute-force (flat index) |
Slow | 100% Accurate | Small datasets |
IVF (ivfflat) |
Fast | ️ Approximate | Large datasets |
Brute-force search is best when the dataset is small (~100K vectors).
IVF indexing is best when the dataset grows large (~millions of vectors) because it avoids scanning every row.
HNSW Indexing in pgvector: A Faster Alternative to IVF
While IVF (Inverted File Indexing) is great for improving search speed in pgvector, another more advanced approach for high-performance vector retrieval is HNSW (Hierarchical Navigable Small World) indexing. HNSW is widely regarded as one of the fastest and most accurate nearest neighbor search algorithms, especially for large datasets.
What is HNSW (Hierarchical Navigable Small World)?
HNSW is a graph-based approach for Approximate Nearest Neighbor (ANN) search. Instead of partitioning data into clusters like IVF, HNSW organizes data into a multi-layered graph where vectors are connected based on similarity.
How HNSW Works Conceptually
-
Graph Construction (Indexing Phase)
- The dataset is structured into a hierarchical graph where similar vectors are linked to one another.
- The top layers of the graph contain a few high-level “entry points” (long-range connections).
- The bottom layers have more detailed, fine-grained connections between closely related vectors.
-
Query Execution (Search Phase)
- A query vector starts at the top layer of the graph.
- It moves through the graph, hopping between increasingly closer nodes at each layer.
- The search eventually lands on the most similar vectors, significantly reducing the number of distance computations.
Why Use HNSW Instead of IVF?
Faster Than IVF → HNSW achieves sub-millisecond query times, even on millions of vectors.
More Accurate → Unlike IVF, which sacrifices accuracy for speed, HNSW maintains high recall while being extremely efficient.
Self-Optimizing Graph → HNSW automatically builds a graph structure that efficiently organizes data for retrieval.
No Need to Tune lists → IVF requires careful tuning of clusters (lists) for performance, but HNSW adapts dynamically.
| Feature | IVF (ivfflat) |
HNSW (hnsw) |
|---|---|---|
| Search Speed | Fast | Very Fast |
| Query Accuracy | ️ Approximate | Higher Accuracy |
| Index Size | Small | Larger (but worth it) |
| Best for | Millions of vectors | Large-scale, low-latency search |
How to Create an HNSW Index in pgvector
You can create an HNSW index using the following SQL command:
-- Create an HNSW index on the embedding column
CREATE INDEX ON bible_embeddings
USING hnsw (embedding vector_l2_ops);
Explanation of Parameters
hnsw→ Tells PostgreSQL to use the HNSW algorithm instead of IVF.vector_l2_ops→ Specifies that L2 (Euclidean distance) should be used for similarity search.
Optional HNSW Parameters for Tuning
HNSW allows customization for better performance:
-- Tune HNSW index parameters
CREATE INDEX ON bible_embeddings
USING hnsw (embedding vector_l2_ops)
WITH (m = 16, ef_construction = 64);
m→ Controls how many links each node has (higherm= better accuracy but slower indexing).ef_construction→ Number of neighbors considered when building the index (higher = better accuracy but longer index time).
For query-time tuning, you can adjust ef_search dynamically:
SET hnsw.ef_search = 50; -- Higher value improves recall but increases latency
When to Use HNSW vs. IVF?
| Use Case | Recommended Index |
|---|---|
| You have a very large dataset (~millions of vectors) | HNSW (faster & more accurate) |
| You need the absolute fastest query times | HNSW |
| Memory efficiency is important | ️ IVF (HNSW uses more RAM) |
| You need fine-tuned speed vs. accuracy trade-offs | HNSW (with ef_search) |
Key Takeaway: If your dataset is large and requires high-speed, high-accuracy vector search, HNSW is the best option in pgvector.
Which Index Should You Choose?
- If you need maximum speed & accuracy → Use HNSW.
- If you need lower memory usage & tunable performance → Use IVF.
- If you have a small dataset (~100K vectors) → Brute-force search (no index) is fine.
Code examples
You can find the code examples for this post and more here
Conclusion
Pgvector turns PostgreSQL into a powerful vector database, enabling semantic search, AI applications, and similarity-based retrieval.
In this guide, we covered:
- Installing PostgreSQL and pgvector
- Creating a vector database and storing embeddings
- Running vector similarity searches with Cosine Similarity & Euclidean Distance
- Using IVF and HNSW indexes to speed up queries on large datasets
Why I Like pgvector I prefer the pgvector solution because it means you can put all of your data in one place the database, keeping everything inside PostgreSQL simplifies management and deployment.
← Back to Blog